[Machine Learning] Random Forest
RandomForest Classifier
|  |
import matplotlib.pyplot as plt
from sklearn.datasets import load_breast_cancer
from sklearn.tree import DecisionTreeClassifier
from sklearn.ensemble import RandomForestClassifier
from sklearn.model_selection import train_test_split
from sklearn import model_selection, svm, metrics
from sklearn.model_selection import GridSearchCV
import numpy as np
import pandas as pd
DecisionTree
cancer = load_breast_cancer()
np.random.seed(9)
X_train, X_test, y_train, y_test = train_test_split(
cancer.data, cancer.target, stratify=cancer.target)
clf = DecisionTreeClassifier()
clf.fit(X_train, y_train)
DecisionTreeClassifier()
print(clf.score(X_test, y_test))
0.916083916083916
X_train.shape, X_test.shape, cancer.data.shape
((426, 30), (143, 30), (569, 30))
feature_importances_:
- In trees, it tells how much each feature contributes to decreasing the weighted impurity.
- in Random Forest, it averages the decrease in impurity over trees.
# 결정 트리를 사용한 경우의 중요 특성
list(zip(cancer.feature_names, clf.feature_importances_.round(4)))[:10]
[('mean radius', 0.0),
('mean texture', 0.0417),
('mean perimeter', 0.0),
('mean area', 0.0),
('mean smoothness', 0.0),
('mean compactness', 0.0),
('mean concavity', 0.0),
('mean concave points', 0.0426),
('mean symmetry', 0.0114),
('mean fractal dimension', 0.0)]
# 정렬해서 상위 10개 검색
df = pd.DataFrame({'feature':cancer.feature_names,'importance':clf.feature_importances_ })
df=df.sort_values('importance', ascending=False)
print(df.head(10))
feature importance
22 worst perimeter 0.694689
27 worst concave points 0.121068
7 mean concave points 0.042647
1 mean texture 0.041720
21 worst texture 0.039639
13 area error 0.017216
20 worst radius 0.017188
15 compactness error 0.012042
8 mean symmetry 0.011405
14 smoothness error 0.002385
x = df.feature
y = df.importance
ypos = np.arange(len(x))
plt.figure(figsize=(10,7))
plt.barh(x, y)
plt.yticks(ypos, x)
plt.xlabel('Importance')
plt.ylabel('Variable')
plt.xlim(0, 1)
plt.ylim(-1, len(x))
plt.show()

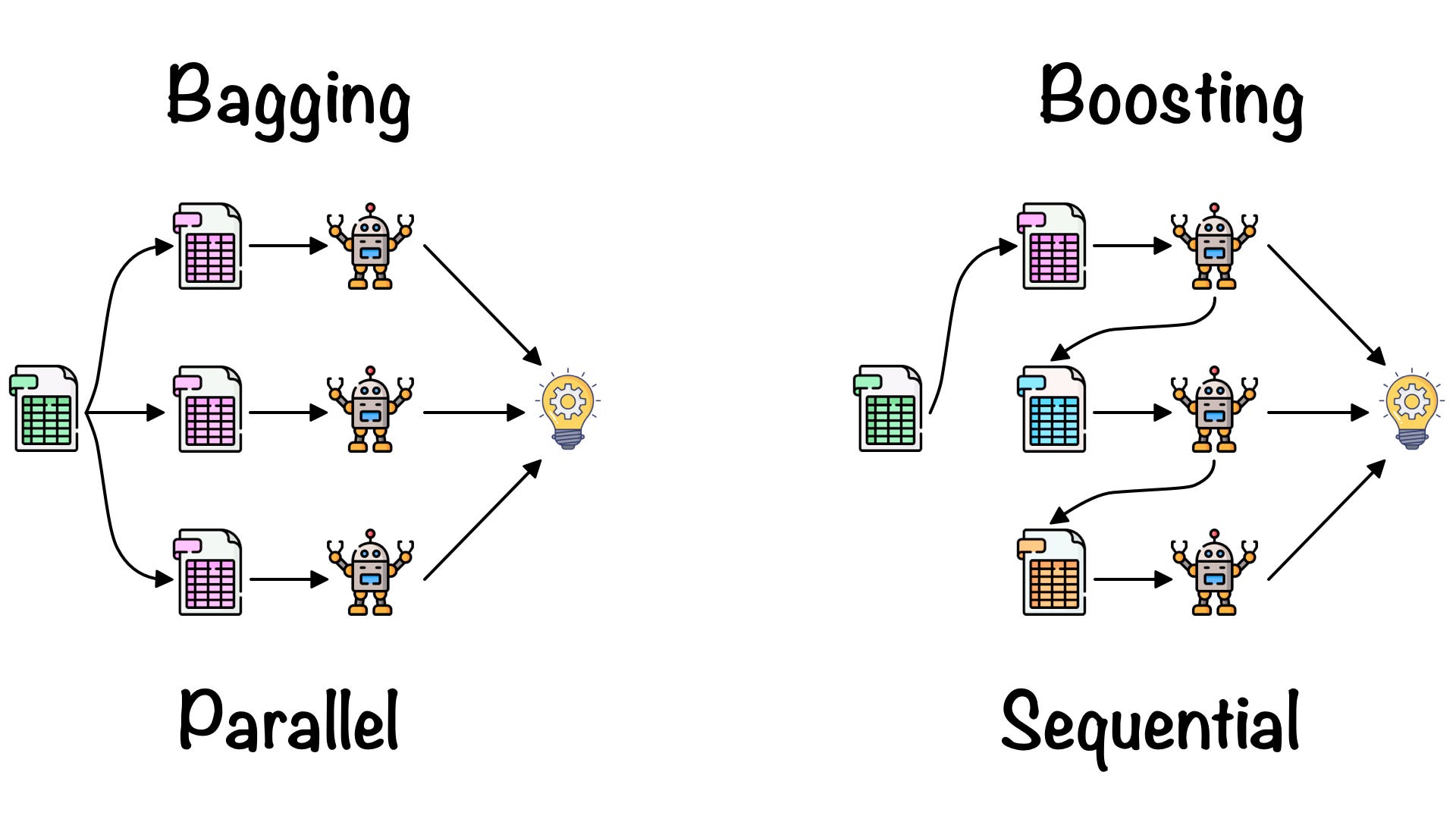
Random Forest (Bagging)
- 중복을 허용한 n개의 샘플을 추출하여 평균을 구하는 작업을 n번 반복
- Overfitting 해소, 일반적인 모델 생성
- Parallel Session
# 랜덤 포레스트를 사용한 경우의 중요 특성
rfc = RandomForestClassifier(n_estimators=500)
rfc.fit(X_train, y_train)
print(rfc.score(X_test, y_test))
df = pd.DataFrame({'feature':cancer.feature_names,'importance':rfc.feature_importances_ })
df=df.sort_values('importance', ascending=False)
x = df.feature
y = df.importance
ypos = np.arange(len(x))
plt.figure(figsize=(10,7))
plt.barh(x, y)
plt.yticks(ypos, x)
plt.xlabel('Importance')
plt.ylabel('Variable')
plt.xlim(0, 1)
plt.ylim(-1, len(x))
plt.show()
0.951048951048951

- 골고루 사용됨을 알 수 있다.
Gradient Boosting (Boosting)
- Bagging은 독립적인 input data를 가지고(복원 추출) 독립적으로 예측하지만, Boosting은 이전 모델이 다음 모델에 영향을 준다.
- 맞추기 어려운 문제를 맞추는 것에 초점을 둔다.
- Serial Session
from sklearn.ensemble import GradientBoostingClassifier
gbc = GradientBoostingClassifier(n_estimators=500)
gbc.fit(X_train, y_train)
print(gbc.score(X_test, y_test))
0.9790209790209791
df = pd.DataFrame({'feature':cancer.feature_names,'importance':gbc.feature_importances_ })
df = df.sort_values('importance', ascending=False)
x = df.feature
y = df.importance
ypos = np.arange(len(x))
plt.figure(figsize=(10,7))
plt.barh(x, y)
plt.yticks(ypos, x)
plt.xlabel('Importance')
plt.ylabel('Variable')
plt.xlim(0, 1)
plt.ylim(-1, len(x))
plt.show()

Leave a comment